My first research project as a Ph.D. student was investigating the differences in the rumen and colon bacterial communities in moose from Vermont. To obtain samples, I needed to secure permission from VT Department of Fish and Wildlife, as moose hunting and moose sample collection are tightly regulated; hunting is only permitted for approximately one week a year, and the licenses are awarded on a lottery system. I was able to contact several permitted hunters for the 2010 season, and passed on sample collection kits to them, complete with gloves, jars, ice packs, a cooler, and instructions with photographs. A total of 14 samples were collected, 8 from the rumen and 6 from the colon – a small sample group in general, but a good-sized one for a wild animal study. If a hunter was successful, they would contact me to collect the samples as soon as possible after the carcass was reported to the state at one of a number of weigh stations. Between dropping off and collecting samples, I drove a total of 1500 miles in about 9 days, and was able to see some beautiful parts of Vermont.
Once back at the lab, the samples were frozen at -80 degrees Celsius to protect the microbial cells and DNA from degradation- I knew I may not get another chance to collect rumen samples from moose. For this project, I used a small amount of each sample and extracted the total mass of nucleic material available – the DNA and some RNA from all the cells present, including microbial, moose, and plant. I then used Polymerase Chain Reaction (PCR) to amplify, or make copes of, just the bacterial DNA. Specifically, I was looking for the 16S rRNA gene, which I would use for amplification. I used a DNA microarray, Phylochip, which had fragments of DNA from known bacteria bound to the chip. Once I applied my sample DNA, if there was a DNA- DNA match, my sample would bind to the chip as well as would fluoresce under ultraviolet light. A sophisticated computer program would read the light signals and interpret then as the presence and rough abundance of that particular species of bacteria.
The most important finding of the study was that the rumen and colon hosted sufficiently different bacterial communities, such that colon or fecal samples were not a good proxy for what was happening in the rumen. While this had been shown once before in sheep, it was not common practice to sample from more than one gastrointestinal location in ruminants, particularly in wild ones, because the cost of sequencing was high and the logistics of sample collection were difficult. Thus, it was common practice to collect fecal samples from wild ruminants and speculate about the rumen microbial communities.
Ishaq, S.L., Wright, A-D.G. 2012. Insight into the bacterial gut microbiome of the North American moose (Alces alces). BMC Microbiology, 12:212. Article
Abstract
Background
The work presented here provides the first intensive insight into the bacterial populations in the digestive tract of the North American moose (Alces alces). Eight free-range moose on natural pasture were sampled, producing eight rumen samples and six colon samples. Second generation (G2) PhyloChips were used to determine the presence of hundreds of operational taxonomic units (OTUs), representing multiple closely related species/strains (>97% identity), found in the rumen and colon of the moose.
Results
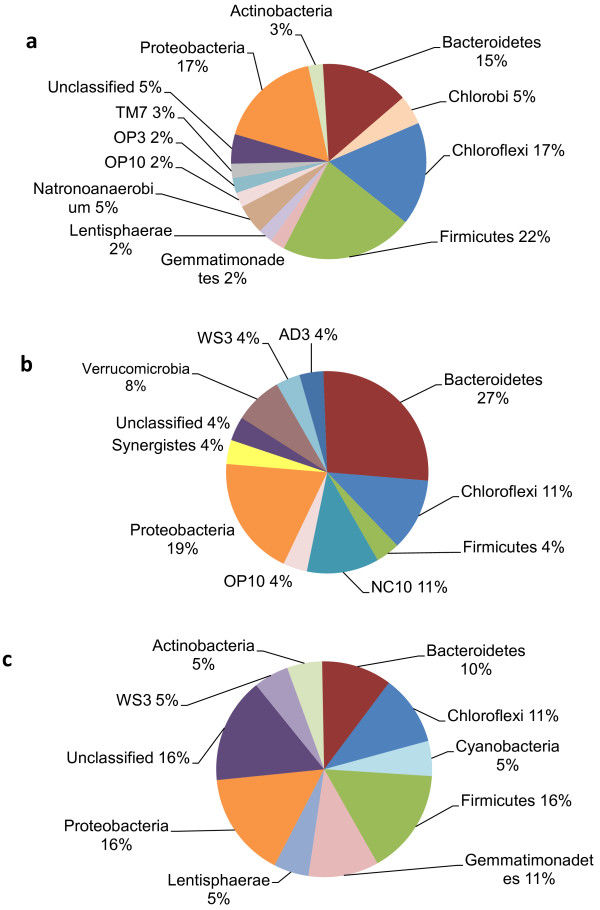
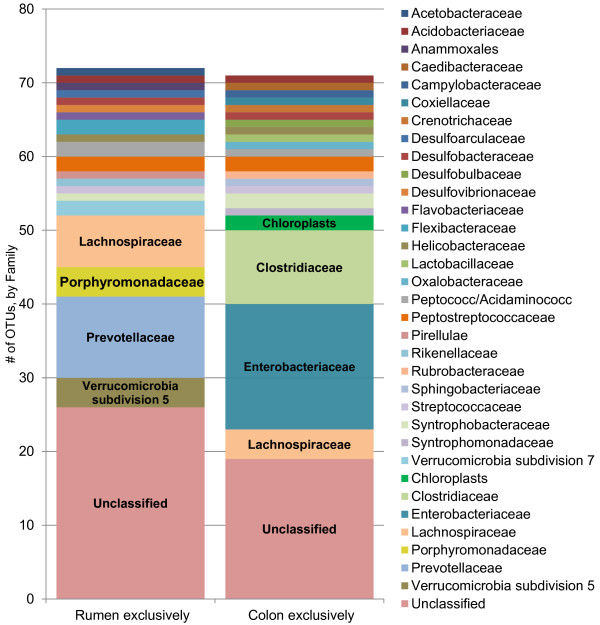
A total of 789 unique OTUs were used for analysis, which passed the fluorescence and the positive fraction thresholds. There were 73 OTUs, representing 21 bacterial families, which were found exclusively in the rumen samples: Lachnospiraceae, Prevotellaceae and several unclassified families, whereas there were 71 OTUs, representing 22 bacterial families, which were found exclusively in the colon samples: Clostridiaceae, Enterobacteriaceae and several unclassified families. Overall, there were 164 OTUs that were found in 100% of the samples. The Firmicutes were the most dominant bacteria phylum in both the rumen and the colon. Microarray data available at ArrayExpress, accession number E-MEXP-3721.
Conclusions
Using PhyloTrac and UniFrac computer software, samples clustered into two distinct groups: rumen and colon, confirming that the rumen and colon are distinct environments. There was an apparent correlation of age to cluster, which will be validated by a larger sample size in future studies, but there were no detectable trends based upon gender.
Keywords