The Ishaq and Li labs at UMaine are delighted to announce that our paper on “Early life exposure to broccoli sprouts confers stronger protection against enterocolitis development in an immunological mouse model of inflammatory bowel disease.” has been published in mSystems!! ASM was kind enough to write a press release about study, found here.
The complete author list, Abstract, and Ackowledgements/Funders portions of the paper can be found at the end of this post. This paper is part of a larger Broccoli project, in which we are evaluating the use of broccoli sprouts in the diet to enlist gut microbes to produce anti-inflammatories as a way to resolve symptoms of Inflammatory Bowel Disease.
The Premise


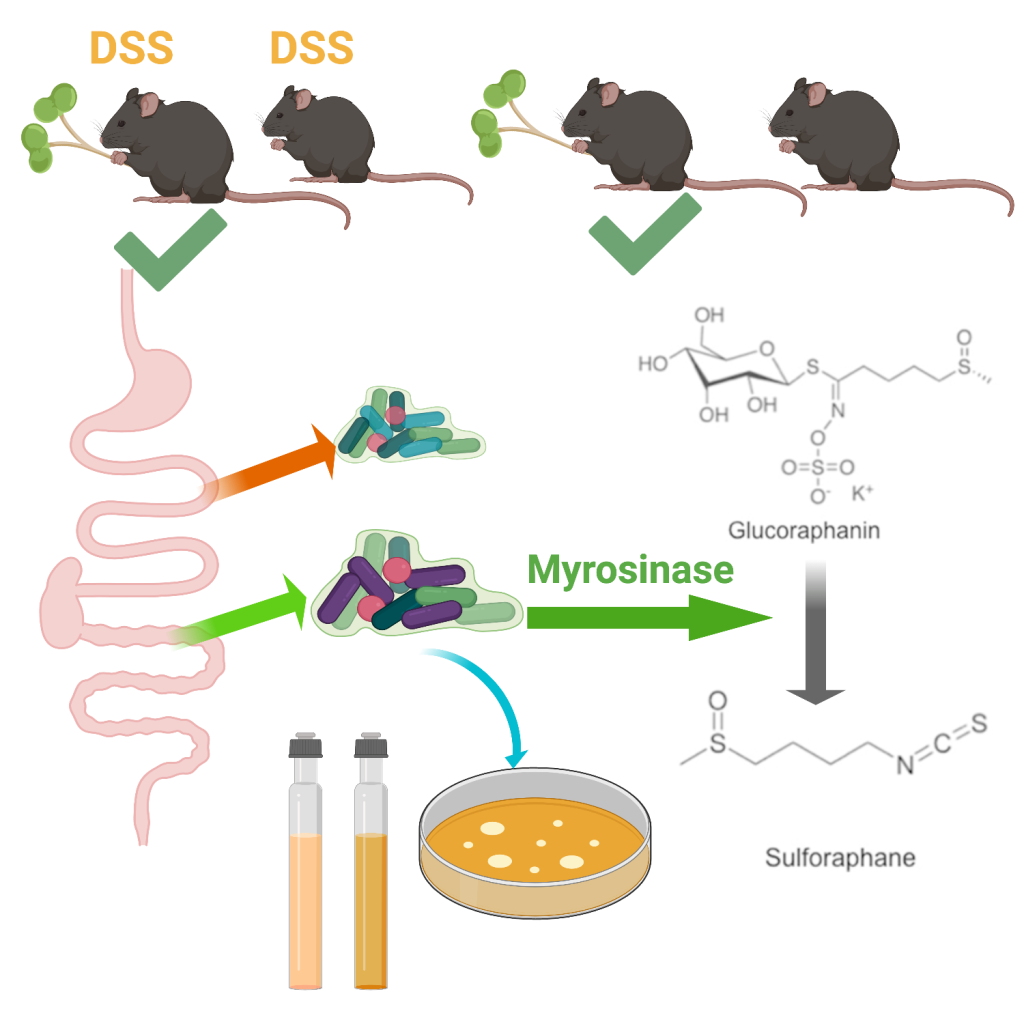
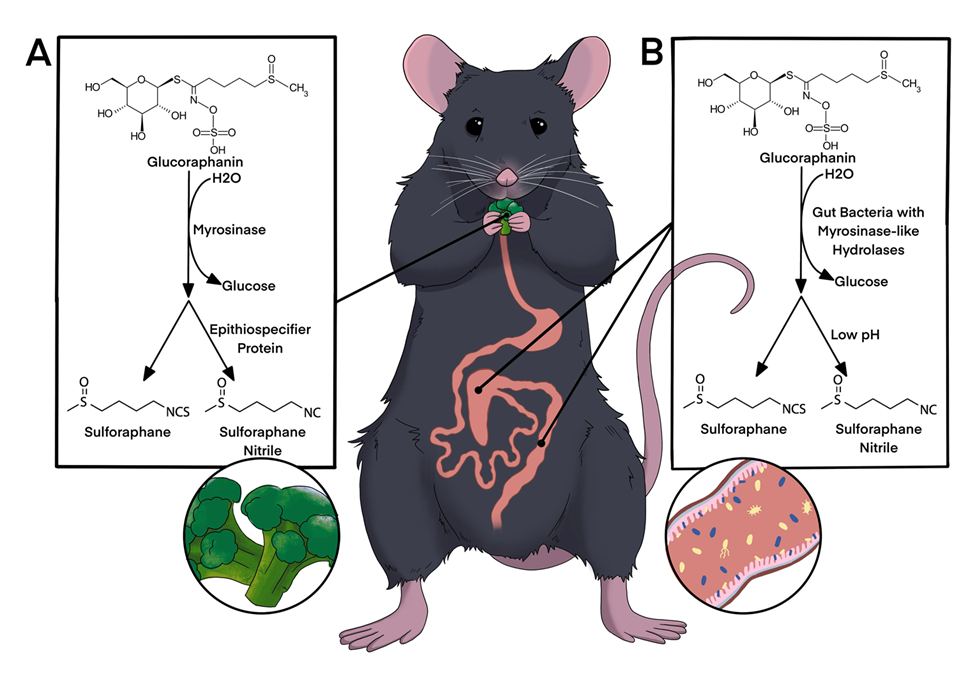
Broccoli sprouts are very high in a compound called glucoraphanin, which is in-active for humans. When glucoraphanin comes in contact with the myrosinase enzyme, also found in the sprouts, it is transformed into sulforaphane, which drives away insect pests but acts as an anti-inflammatory in people!
If you eat raw sprouts, most of this conversion happens when you cut or chew the sprouts, and that anti-inflammatory will get absorbed in your stomach. If you steam or cook the sprouts, you can inactivate the enzyme and leave the glucoraphanin compound alone. Some of your gut microbes are able to use glucoraphanin, and produce the anti-inflammatory sulforaphane right in your gut! We are trying to understand how and when this works, so we can use it to reduce symptoms of Inflammatory Bowel Disease.

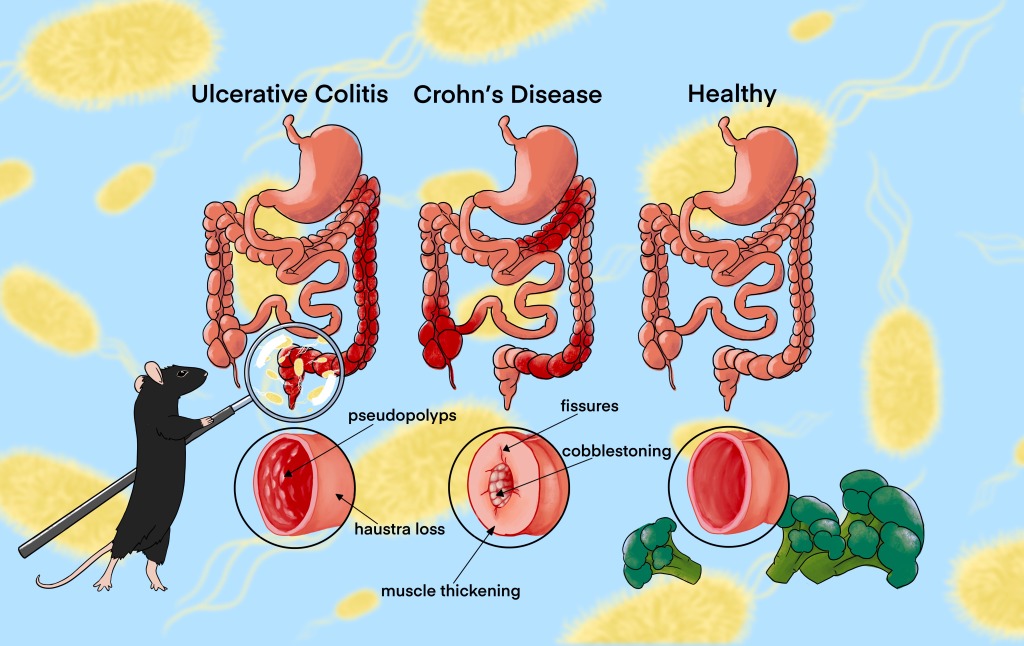
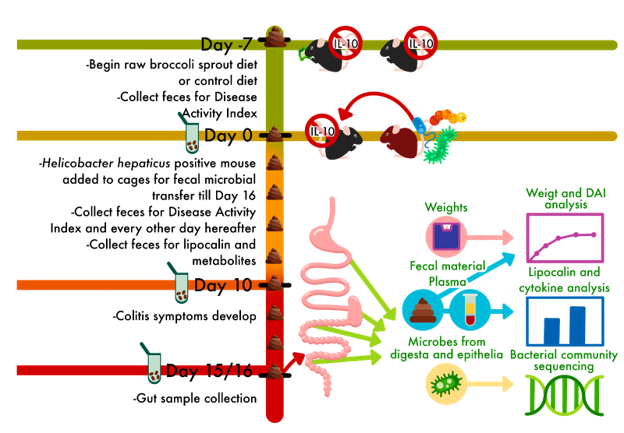
The mice in this trial are used to mimic Crohn’s Disease, which is one of the main ways that Inflammatory Bowel Diseases may be classified. Crohn’s Disease is complictaed, and involves an over-active immune response to gut microbes. This is replicated in mice that are bred to lack the genes in the DNA to make interleukin-10 (IL-10). IL-10 is an immune factor that can be used to calm the immune system and tolerate microbes which are not causing harm. Without IL-10, these mice over-react to the presence of bacteria, even those which are not causing harm, and this creates symptoms similar to Crohn’s in people.
We used two age groups of mice, and in each group, half ate a mouse chow (control) diet and half ate the mouse chow with 10% of the chow replaced by raw broccoli sprouts. Crohn’s often develops in childhood and adolescence, so our two age groups of mice reflect the juvenile stage (4-5 weeks old) and the adolescence stage (5-6 weeks old) of symptom onset. After wo weeks of symptoms, we sacrificed the mice and collected as much information as we could.
The Team
The mice, their care during the experiment, and sample collection for this project was graciously provided by University of Vermont researchers Gary Mawe and Brigitte Lavoie, and then-grad-student-now-medical-student Molly Hurd, in 2021. The SUNY Bingamton team, Tao Zhang and Allesandra Stratigakis, processed metabolite and cytokine samples and analyzed those data. The UMaine team (pictured below and led by Sue Ishaq and Yanyan Li) processed and analyzed data from different locations of gut tissue for histolgy and sequencing of bacterial communities, as well as analyzing those data, and took the lead on writing the paper.







The Health Benefits were most obvious in the younger mice
The mice that were eating the broccoli sprouts in their chow and did much better than the control group who ate only mouse chow when symptoms of Crohn’s Disease were induced — and we found something really interesting… The diet worked really well in the younger mice and reduced their symtpoms of inflammation and illness for almost every metric we studied. The older, adolecent mice got some benefit from eating the raw broccoli sprouts, but not nearly as much as the younger mice! Those graphs are shown in the paper.
The Gut Microbes were most changed in the younger mice
Bacterial richness (the number of different types of bacteria present) was increased, but only in younger mice consuming a 10% raw sprout diet, which is useful because pediatric Crohn’s patients usually have fewer types of bacteria present in their gut.

Younger mice consuming broccoli sprouts also had more types of bacteria that are known to convert glucoraphanin into sulforophane, and they had more of the genes needed to do it. Crohn’s patients usually have fewer of these types of bacteria, which are also known to provide other health benefits.
The Next Steps
We are currently working on replicating and expanding this project to include more age groups, so we can understand how different diet preparations of broccoli sprouts impact immune systems and gut microbiota at different developmental periods of life. We are also really interested in understanding how sex in mice, and gender in humans, plays a role in how immune systems and microbial communities develop during a critical phase of life. We have some initial data to suggest that male and female mice respond to different diets and at differnt ages, but we aren’t sure why yet.
We hope to expand our work with people to study how these diets work in the real world, and how we can tailor diet and cooking preparations of sprouts to best meet the needs of people of different ages, health statuses, and tastes.
The paper
Early life exposure to broccoli sprouts confers stronger protection against enterocolitis development in an immunological mouse model of inflammatory bowel disease.
Lola Holcomb1$, Johanna M. Holman2$, Molly Hurd3, Brigitte Lavoie3, Louisa Colucci4, Benjamin Hunt5, Timothy Hunt5, Marissa Kinney2, Jahnavi Pathak1, Gary M. Mawe3,Peter L. Moses3,6, Emma Perry7, Allesandra Stratigakis8, Tao Zhang8, Grace Chen9, Suzanne L. Ishaq1*, Yanyan Li1*
1 Graduate School of Biomedical Sciences and Engineering, University of Maine, Orono, Maine, USA 04469. 2 School of Food and Agriculture, University of Maine, Orono, Maine, USA 04469. 3 Larner College of Medicine, University of Vermont, Burlington, Vermont, USA 05401. 4 Department of Biology, Husson University, Bangor, Maine, USA 04401. 5 Department of Biology, University of Maine, Orono, Maine, USA 04469. 6 Finch Therapeutics, Somerville, Massachusetts, USA 02143. 7 Electron Microscopy Laboratory, University of Maine, Orono, Maine, USA 04469. 8 School of Pharmacy and Pharmaceutical Sciences, SUNY Binghamton University, Johnson City, New York, USA 13790. 9 Department of Internal Medicine, University of Michigan Medical School, Ann Arbor, Michigan, USA 48109
$ these authors contributed equally.
Keywords: Crohn’s Disease, cruciferous vegetables, sulforaphane, glucoraphanin, gut microbiota, dietary bioactives, 16S rDNA, interleukin-10 knockout
Abstract
Crohn’s Disease (CD) is a presentation of Inflammatory Bowel Disease (IBD) that manifests in childhood and adolescence, and involves chronic and severe enterocolitis, immune and gut microbial dysregulation, and other complications. Diet and gut-microbiota-produced metabolites are sources of anti-inflammatories which could ameliorate symptoms. However, questions remain on how IBD influences biogeographic patterns of microbial location and function in the gut, how early life transitional gut communities are affected by IBD and diet interventions, and how disruption to biogeography alters disease mediation by diet components or microbial metabolites. Many studies on diet and IBD use a chemically induced ulcerative colitis model, despite the availability of an immune-modulated CD model. Interleukin-10-knockout (IL-10-KO) mice on a C57BL/6 background, beginning at age 4 or 7 weeks, were fed a control diet or one containing 10% (w/w) raw broccoli sprouts, which was high in the sprout-sourced anti-inflammatory sulforaphane. Diets began 7 days prior to, and for 2 weeks after inoculation with Helicobacter hepaticus, which triggers Crohn’s-like symptoms in these immune-impaired mice. The broccoli sprout diet increased sulforaphane in plasma; decreased weight stagnation, fecal blood, and diarrhea associated; and increased microbiota richness in the gut, especially in younger mice. Sprout diets resulted in some anatomically specific bacteria in younger mice, and reduced the prevalence and abundance of pathobiont bacteria which trigger inflammation in the IL-10-KO mouse, e.g., Escherichia coli and Helicobacter. Overall, the IL-10-KO mouse model is responsive to a raw broccoli sprout diet and represents an opportunity for more diet-host-microbiome research.
Importance
To our knowledge, IL-10-KO mice have not previously been used to investigate the interactions of host, microbiota, and broccoli, broccoli sprouts, or broccoli bioactives in resolving symptoms of CD. We showed that a diet containing 10% raw broccoli sprouts increased the plasma concentration of the anti-inflammatory compound sulforaphane, and protected mice to varying degrees against disease symptoms, including weight loss or stagnation, fecal blood, and diarrhea. Younger mice responded more strongly to the diet, further reducing symptoms, as well as increased gut bacterial richness, increased bacterial community similarity to each other, and more location-specific communities than older mice on the diet intervention. Crohn’s Disease disrupts the lives of patients, and requires people to alter dietary and lifestyle habits to manage symptoms. The current medical treatment is expensive with significant side effects, and a dietary intervention represents an affordable, accessible, and simple strategy to reduce the burden of symptoms.
Acknowledgements: This project was supported by the USDA National Institute of Food and Agriculture through the Maine Agricultural & Forest Experiment Station: Hatch Project Numbers ME022102 and ME022329 (Ishaq) and ME022303 (Li); the USDA-NIFA-AFRI Foundational Program [Li and Chen; USDA/NIFA 2018-67017-27520/2018-67017-36797]; and the National Institute of Health [Li and Ishaq; NIH/NIDDK 1R15DK133826-01] which supported Marissa Kinney, Timothy Hunt, and Benjamin Hunt. Johanna Holman was supported by ME0-22303 (Li), and Lola Holcomb was supported by US National Science Foundation One Health and the Environment (OG&E): Convergence of Social and Biological Sciences NRT program grant DGE-1922560, and the UMaine Graduate School of Biomedical Science and Engineering.